Computational BioMedicine
& RNA Biology
Computational BioMedicine
& RNA Biology
In the Computational BioMedicine lab, we develop integrative workflows combining various computational disciplines with experimentation to address questions around non-coding RNAs, post-transcriptional gene regulation and cancer biology. Moreover, we develop multi-omics data analysis workflows to investigate patterns of alternative splicing in normal biology and in various cancers.
We have identified intron retention as a well conserved form of alternative splicing that mediates cell-specific gene regulation. Aberrant intron retention has been described in multiple human cancers. Using machine learning, mathematical modelling, and molecular dynamics simulations we investigate the role of this and other forms of post-transcriptional gene regulation in cancer pathophysiology and therapy resistance.
Projects
Alternative splicing and the epigenome in Chronic Myeloid Leukaemia
Mechanisms of intron retention regulation in chronic myeloid leukemia
In this project, we are investigating gene regulatory processes in leukaemia. The third biggest cause of cancer death in all Australians is blood cancers (leukaemia), which are diagnosed 35 times each day. Using a multi-omics approach, we examine alternative splicing and epigenetic changes blood samples from chronic myeloid leukaemia (CML) patients before and after treatment with tyrosine kinase inhibitors.
Find out more: Schmitz et al, Cancers 2020
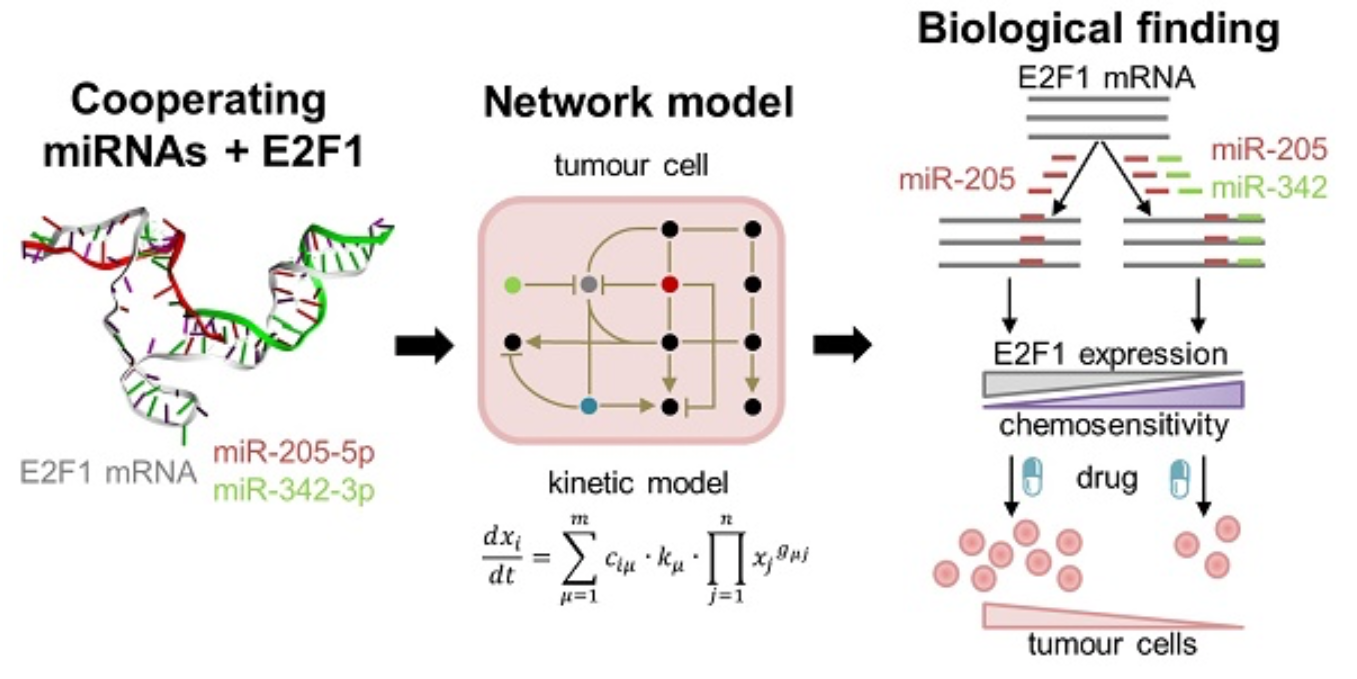
Cooperating microRNAs for cancer therapy
Pairs of cooperating microRNAs used as RNA therapeutics to reduce chemoresistance
In this project, we use a systems medicine approach to find new avenues for overcoming chemotherapy resistance in aggressive tumour cells. Our results suggest that pairs of cooperating microRNAs could be used as potential RNA therapeutics to reduce E2F1-related chemoresistance.
Find out more: Lai X, Gupta SK, Schmitz U et al, Theranostics 2018
Cross-talk between post-transcriptional gene regulation mechanisms
Intron retention-mediated microRNA sponge mechanism
Combining computational predictions and in vitro assays we elucidate the cross-talk between post-transcriptional gene regulation mechanisms. More specifically, we focus in interactions between microRNA molecules and intron-retaining mRNA transcripts and try to identify networks of interconnected gene regulation. Using a systems biology approach we will model competitive post-transcriptional gene regulation through iterative cycles of time course experiments and model simulations.
Intron retention regulation in haematopoietic cells
Model of intron retention regulation in haematopoietic cells
There is growing evidence that alternative splicing, including IR, is regulated on at least two levels: locally, through a network of interacting trans-acting splicing regulators with cis-acting regulatory elements, and globally, through the chromatin structure. In this project, we explore a global regulation of IR in haematopoietic cells – we use statistical analysis to better understand IR regulation using information derived from the epigenetic factors that govern chromatin organisation, namely nucleosome assembly, DNA methylation and histone modifications. The main challenge here is to integrate multiple layers of ‘-omics’ into a single computational model which produces biologically interpretable results.
The role of zinc finger proteins in alternative splicing regulation
CTCF and ZRANB2 are two zinc finger proteins that have been recently linked to alternative splicing. We carried out transcriptomic and proteomic analyses using Ctcf and Zranb2 transgenic mice to identify alternative splicing events and interacting splicing factor proteins across different mouse tissues.
Find out more: Alharbi et al, RNA Biology 2020
Model of Ctcf as modulator of alternative splicing
Selected publications from the Computation BioMedicine lab
Widespread Aberrant Alternative Splicing despite Molecular Remission in Chronic Myeloid Leukaemia Patients. Schmitz U, Shah JS, Dhungel BP, Monteuuis G, Luu PL, Petrova V, Metierre C, Nair SS, Bailey CG, Saunders VA, Turhan AG, White DL, Branford S, Clark SJ, Hughes TP, Wong JJ, Rasko JEJ. (2020) Cancers (Basel). 11;12(12):E3738
The changing paradigm of intron retention: Regulation, Ramifications and Recipes. Monteuuis G, Wong JJ-L, Bailey CG, Schmitz U*, Rasko JEJ* (2019) Nucleic Acids Research, Vol 47, Issue 22:16, 11497–11513. *co-corresponding author
MiR-205-5p and miR-342-3p cooperate in the repression of the E2F1 transcription factor in the context of anticancer chemotherapy resistance. Lai X*, Gupta SK*, Schmitz U*, Marquard S, Knoll S, Wolkenhauer O, Puetzer B, Vera J (2018) Theranostics, 8(4): 1106-1120. *co-first author
Intron retention enhances gene regulatory complexity in vertebrates. Schmitz U, Pinello N, Jia F, Alasmari S, Ritchie W, Keightley M-C, Shini S, Lieschke GJ, Wong JJ-L, Rasko JEJ (2017) Genome Biology 18.1: 216.
Cooperative gene regulation by miRNA pairs and their identification using a computational workflow. Schmitz U, Lai X, Winter F, Wolkenhauer O, Vera J, Gupta S (2014)Nucleic Acid Research, 42, 12, p. 7539- 7552, doi: 10.1093/nar/gku465.